Recap
Alright, so last time, I hit a wall with memory allocation when attempting to analyze ambient-temperature crabs.
At first, I tried to get around this issue by just heading into UW and using the computers there. That partially solved the issue, but not entirely. At one stage in the pipeline, you set a bar that determines what genes should be discarded - the original recommendation is genes where counts are below 10 in 90% of samples, but since I had only 9 samples, just changed it to counts below 10 in all samples. Problem: that crashes the lab computers entirely, and even upping the bar to 15 takes practically an entire day to run.
So as an alternative, I started using RStudio on Mox, following the guidelines figured out here. That partially worked, but it still takes a really long time to run 10 samples, so I decided to get some preliminary results by setting the bar at rejecting genes with counts below 30 in all samples. That ran smoothly, and so here’s what I have! [NOTE: later realized I made errors in all of these - you shouldn’t run WGCNA with less than 9 samples! I’ve linked to some updated examples involving all crabs, just so you can visualize some possible results]

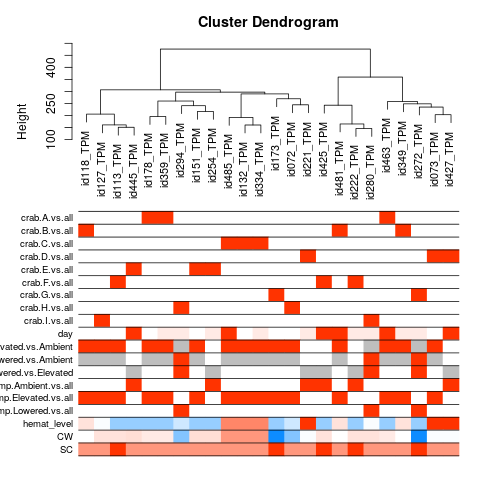
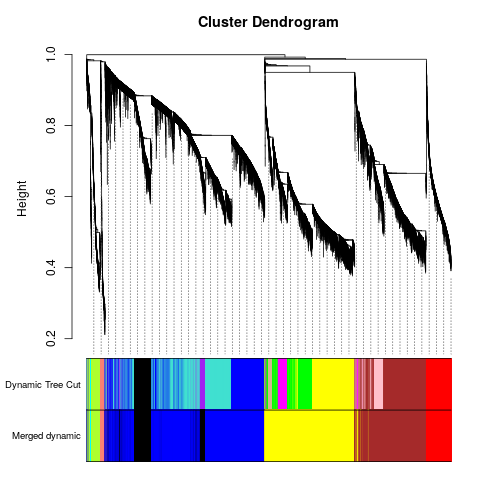
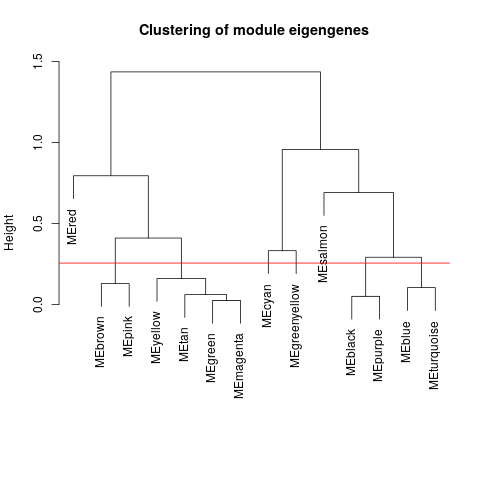
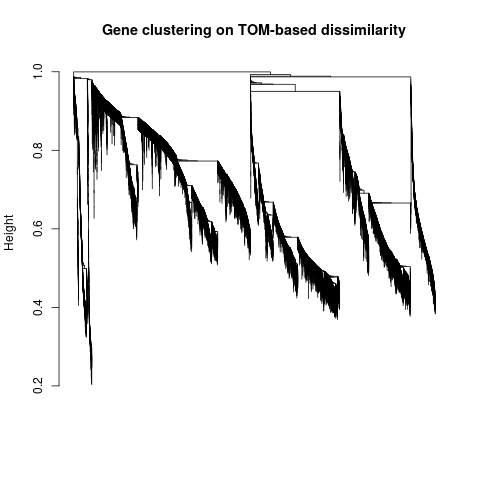
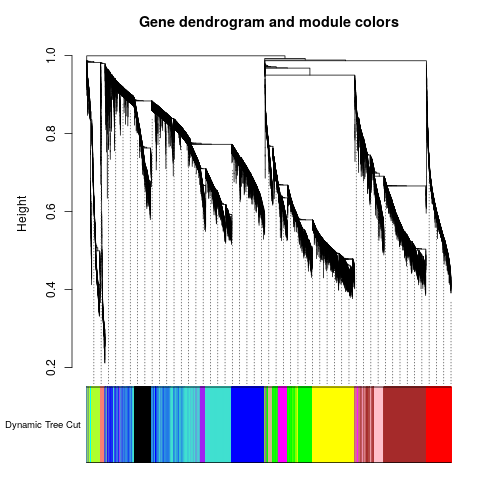
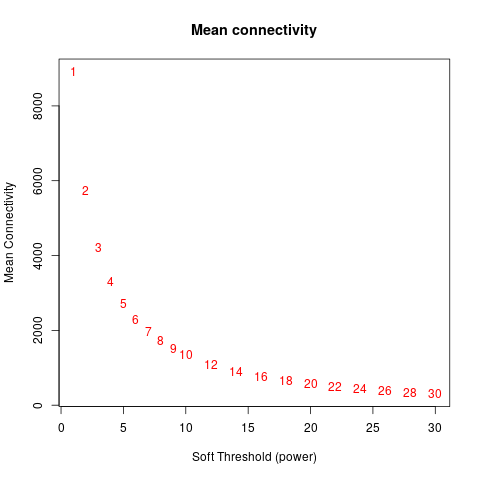
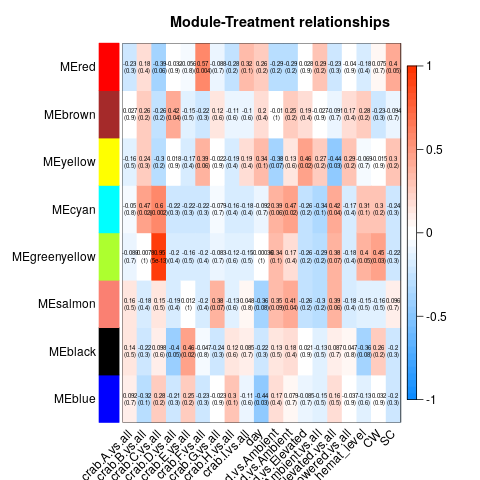
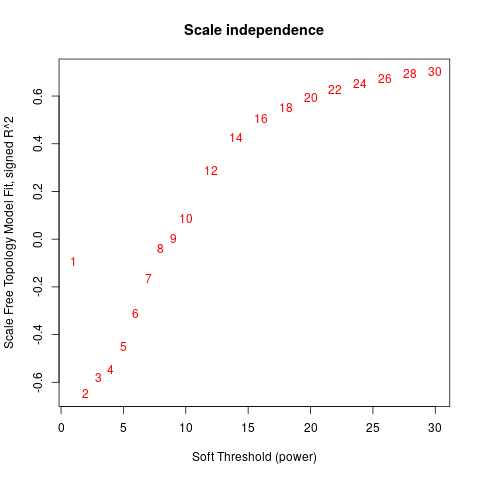
Specific gene lists for each eigengene
The heat map is particularly interesting - seems to not show any significant relationships with day (if we set the p-value bar at 0.05), but some interesting relationships with individual crab.
Anyway, I think I need some help interpreting these results and deciding my next steps to take. Some possible next steps:
-
Recreate this with Day 0/2 Elevated crabs
-
Recreate this with Day 2 Ambient/Day 2 Elevated crabs
-
Continue by performing GO-MWU on each WGCNA module
I’ll post this as a lab discussion too, hopefully I get some good tips on where to go next!